We will work with the traditional “five frequency” model for solute diffusion in a face-centered cubic (FCC) lattice. A relatively large database of solute barriers was published by the Dane Morgan group recently; paper with the public database which we will use for our computation.
import pandas as pd
import numpy as np
from numpy.random import default_rng
rng = default_rng()
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.constants
# grab Boltzmann's constant; we'll need it later
kBoltzmann = scipy.constants.physical_constants['Boltzmann constant in eV/K'][0]The datafile we need is an Excel spreadsheet, available at https://ndownloader.figshare.com/files/12753815. In addition, you should investigate the structure of the database at https://figshare.com/articles/dataset/DFT_dilute_solute_diffusion_in_Al_Cu_Ni_Pd_Pt_and_Mg/1546772
We use sheet_name=None to get all of the sheets; the first sheet is the host information, followed by different solutes.
Some information about what we’re going to see:
A_shift and energy shift E_shift that they apply to the diffusivity data, of the form \(A_\text{shift}\exp(-E_\text{shift}/k_\text{B}T)\).diffusionDB = pd.read_excel('https://ndownloader.figshare.com/files/12753815',
sheet_name=None)You’ll want to inspect the content of the sheets to understand what the data looks like.
diffusionDB.keys()dict_keys(['Host Information', 'Al-X', 'Cu-X', 'Ni-X', 'Pd-X', 'Pt-X', 'Mg-X', 'W-X', 'Mo-X', 'Fe-X', 'Au-X', 'Ca-X', 'Ir-X', 'Pb-X', 'Ag-X', 'Zr-X'])diffusionDB['Host Information']| Host element name | Al | Cu | Ni | Pd | Pt | Mg | W | Mo | Fe | Au | Ca | Ir | Pb | Ag | Unnamed: 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Host crystal structure | FCC | FCC | FCC | FCC | FCC | HCP | BCC | BCC | BCC | FCC | FCC | FCC | FCC | FCC | NaN |
| 1 | Host melting temperature [K] | 933 | 1358 | 1728 | 1828 | 2041 | 923 | 3695 | 2896 | 1811 | 1337 | 1115 | 2719 | 600 | 1234 | NaN |
| 2 | Host vacancy formation energy [eV] | 0.4847 | 0.9628 | 1.39014 | 1.1365 | 0.6112 | 0.7978 | 3.052665 | 2.936949 | 2.22277 | 0.4097 | 1.131886 | 1.582394 | 0.416922 | 0.78365 | NaN |
| 3 | Host lattice constant, a [angstrom] | 4.039094 | 3.633779 | 3.507861 | 3.933174 | 3.962852 | 3.193777 | 3.18834 | 3.157333 | 2.82491 | 4.1565 | 5.504164 | 3.87636 | 5.015953 | 4.142557 | NaN |
| 4 | Host lattice constant, c [angstrom] | NaN | NaN | NaN | NaN | NaN | 5.175814 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 5 | Host self-diffusion correction shift, A_shift | 12 | 80 | 500 | 20 | 20 | 240 | 800 | 2400 | 1000 | 60 | 1260 | 85 | 240 | 55 | NaN |
| 6 | Host self-diffusion correction shift, E_shift … | 0.2 | 0.4 | 0.47 | 0.55 | 0.85 | 0.2 | 1.3 | 1 | 0.08 | 0.85 | 0.09 | 0.42 | 0.1 | 0.43 | NaN |
| 7 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 8 | Additional Information | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | Diffusion values for Fe-X are given for the al… | NaN | NaN | NaN | NaN | NaN | NaN |
| 9 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | The paramagnetic D0 and Q are given here, the … | NaN | NaN | NaN | NaN | NaN | NaN |
| 10 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | D_BCC(T) = D0_para * exp [-Q_para(1+αs^2) / … | NaN | NaN | NaN | NaN | NaN | NaN |
| 11 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | Where α=0.156 and s is the temperature depende… | NaN | NaN | NaN | NaN | NaN | NaN |
Let’s work with Aluminum, and examine the information available for the host:
host_element = 'Al'
diffusionDB['Host Information'][host_element]0 FCC
1 933
2 0.4847
3 4.039094
4 NaN
5 12
6 0.2
7 NaN
8 NaN
9 NaN
10 NaN
11 NaN
Name: Al, dtype: objectWe’ll need to grab some of this information for later:
a0 = diffusionDB['Host Information'][host_element][3]
Evform = diffusionDB['Host Information'][host_element][2]
A_shift, E_shift = diffusionDB['Host Information'][host_element][5], diffusionDB['Host Information'][host_element][6]We can investigate the solute data sheet:
solute_elements = host_element + '-X'And we select one species (Ag) to consider:
soluteZ = 47
diffusionDB[solute_elements][soluteZ]0 Ag
1 0.58143
2 0.57299
3 0.45938
4 0.59649
5 0.52671
6 4.224243
7 4.202993
8 1.322896
9 4.226324
10 4.17018
11 0.040325
12 1.141065
13 NaN
14 NaN
15 NaN
16 NaN
Name: 47, dtype: objectLet’s grab data for that particular solute:
df = diffusionDB[solute_elements][soluteZ]
solute = df[0]
barriers = df[1:6].astype('float').to_numpy()
attempt_freq = df[6:11].astype('float').to_numpy()
D0_diff, Q_diff = df[11], db[12]
print(barriers)
print(attempt_freq)
print(solute, D0_diff, Q_diff)[0.58143 0.57299 0.45938 0.59649 0.52671]
[4.22424298 4.20299328 1.32289592 4.22632423 4.17017994]
Ag 0.0403254943845161 1.14106512869029To compute solute diffusivity, we need to know how to transform this data into diffusivity. There are a few steps involved:
The former is done using the Arrhenius relation; for activation barrier \(Q\) and attempt frequency \(\nu\) at temperature \(T\), the rate \(w\) is \[w = \nu \exp\left(-\frac{Q}{k_\text{B} T}\right)\] where \(k_\text{B}\) is Boltzmann’s constant.
The latter is done using the closed-form expression for the five-frequency model. The rates are
w0: vacancy hop rate; independent of the solute, only dependent on the host.w1: vacancy swing rate: jump rate for a vacancy next to a solute, jumping to another neighbor site of the solutew2: solute exchange rate: jump rate for a solute to jump into the vacant sitew3: vacancy escape rate: jump rate for a vacancy next to a solute, jumping away from the solutew4: vacancy capture rate: jump rate for a vacancy away from a solute, jumping next to a soluteNote that the ratio \(p=w_4/w_3\) is the enhancement probability for a vacancy to be found next to a solute; \(p>1\) means that vacancies are attracted to the solute, and \(p<1\) means that vacancies are repelled by solutes. The derivation of the five frequency diffusivity is a bit complex. Note that when \(w_2\) is very small, the diffusivity is proportional to \(p\cdot w_2\), but when \(w_2\) is very larger, the diffusivity is proportional to \(p(w_1 + w_3 F/2)\) where \(F\) is a correlation factor. The expression for the diffusivity is: \[D = a_0^2 c_\text{v} f(w_0, w_1, w_2, w_3, w_4)\] for lattice constant \(a_0\), equilibrium vacancy concentration \(c_\text{v} = \exp(-E^\text{form}_\text{v}/k_\text{B}T)\) and five-frequency function \(f\).
def fivefreq(w0, w1, w2, w3, w4):
"""The solute diffusion correlation factor in the 5-freq. model"""
b = w4 / w0
F7 = 7. - b * (1338.1 + b * (924.3 + b * (180.3 + b * 10.))) / \
(435.3 + b * (596. + b * (253.3 + b * (40.1 + b * 2.))))
p = w4 / w3
return p * w2 * (2. * w1 + w3 * F7) / (2. * w2 + 2. * w1 + w3 * F7)def Arrhenius(Q, nu, T):
"""Takes activation barrier Q in eV, nu in THz, T in K, and returns a rate in THz"""
return nu*np.exp(-Q/(kBoltzmann*T))Arrhenius(1.0, 1.0e12, 300)1.5875937551666036e-05Now, try comparing the Arrhenius fit for the solute data against the direct five-frequency calculation:
T = 600
D = Arrhenius(Q_diff, D0_diff, T)
D1.049674050545576e-11cV = Arrhenius(Evform, 1., T)
w = Arrhenius(barriers, 1e12*attempt_freq, T)
(a0*1e-8)**2 * cV * fivefreq(*w) * Arrhenius(E_shift, A_shift, T)1.0718133901395067e-11We can apply an approach similar to that used in Uncertainty quantification of solute transport coefficients: with an empirical estimate of the covariance matrix, we can generate sample activation barriers, and determine the distribution of predictions of diffusivity.
Our empirical correlation matrix for this model is below. It is approximate, with a form similar to that found for solutes in HCP Mg. In this case, it is a \(6\times 6\) matrix, with the first entry for the vacancy formation energy, followed by the 5 rates. There is an anticorrelation between the rates w3 and w4, along with reduced correlation with the vacancy formation energy.
barrier_covariance = 0.003*np.eye(6) + 0.003*np.ones((6,6))
barrier_covariance[4,5] = -0.001
barrier_covariance[5,4] = -0.001
barrier_covariance[0,5] = 0.
barrier_covariance[5,0] = 0.
barrier_covariancearray([[ 0.006, 0.003, 0.003, 0.003, 0.003, 0. ],
[ 0.003, 0.006, 0.003, 0.003, 0.003, 0.003],
[ 0.003, 0.003, 0.006, 0.003, 0.003, 0.003],
[ 0.003, 0.003, 0.003, 0.006, 0.003, 0.003],
[ 0.003, 0.003, 0.003, 0.003, 0.006, -0.001],
[ 0. , 0.003, 0.003, 0.003, -0.001, 0.006]])np.linalg.eigh(barrier_covariance)(array([8.00831974e-05, 3.00000000e-03, 3.00000000e-03, 3.00000000e-03,
7.86296696e-03, 1.90569498e-02]),
array([[-2.73008069e-01, -7.51468311e-01, -1.59955093e-01,
2.03196193e-01, 3.66220423e-01, -3.99712708e-01],
[ 3.38527167e-01, 2.80877441e-01, -1.31172585e-02,
7.69407845e-01, -9.78281429e-02, -4.52522243e-01],
[ 3.38527167e-01, 8.47034804e-02, -6.65704257e-01,
-4.69803996e-01, -9.78281429e-02, -4.52522243e-01],
[ 3.38527167e-01, -1.77713844e-01, 7.18810288e-01,
-3.50402898e-01, -9.78281429e-02, -4.52522243e-01],
[-4.76853148e-01, 5.63601233e-01, 1.19966319e-01,
-1.52397145e-01, 5.20903278e-01, -3.82109529e-01],
[-5.95210670e-01, -2.77555756e-17, 1.11022302e-16,
-1.11022302e-16, -7.52217615e-01, -2.82653353e-01]]))To generate our random samples, we use the multivariate_normal function. Here is how we can use it to construct a distribution of diffusivities (here, stored as the log of the diffusivities) by generating samples. Note: we need to prepend the vacancy formation energy to the barrier list so that the energies that come out are a vector [Evform, w0barrier, w1barrier, w2barrier, w3barrier, w4barrier].
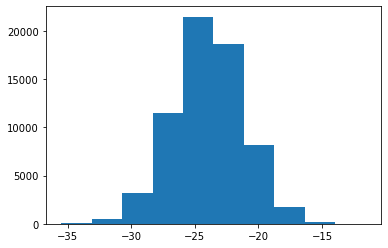
logD = []
T = 600
Nsamples = 2**16
for ene in rng.multivariate_normal(mean=np.insert(barriers, 0, Evform),
cov=barrier_covariance,
size=Nsamples):
cV = Arrhenius(ene[0], 1., T)
w = Arrhenius(ene[1:], 1e12*attempt_freq, T)
D = (a0*1e-8)**2 * cV * fivefreq(*w)
logD.append(np.log(D))
logD = np.array(logD)np.mean(logD), np.std(logD)(-23.936257895471147, 2.7688013872446393)plt.hist(logD);
With this in mind, we want to investigate some questions: